GSoC Mentor Summit 2019 ☀️
Just got to the airport in Munich after a successful Google Summer of Code (GSoC) Mentor Summit. This is a yearly event where all open source organizations that participated in GSoC come together. There are a few scheduled sessions but the rest follows the unconference formula:
Really like the unconference idea: give participants a bunch of rooms and time slots and let them organize their own sessions. Participants know what will be useful to them! Doing an Open Source Bioinformatics session at 11am tomorrow (top left) pic.twitter.com/OK0c9p1bTP
— 🔧 Ino de Bruijn 🧬 (@inodb) October 18, 2019
I was there having a blast representing the cBioPortal organization together with Angelica. This is us:

The cBioPortal organization had 6 students coding over the summer, working on various projects related to the Open Source cBioPortal for Cancer Genomics website. The majority of the contributors are at Memorial Sloan Kettering Cancer Center in New York but the users, a combination of clinicians and researchers, live all over the world and so do its contributors. For more information about cBioPortal see here. For a complete overview of the student projects see the cBioPortal GSoC wiki.
Before I dive into the GSoC mentor summit and elaborate why it was useful to go other than experiencing the great joys of drinking giant beers and eating wild deer schnitzel; I need to share some experiences from Munich. I won’t be offended if you prefer to skip the next paragraph.
Munich 🛴
I loved that the summit was in Munich this year. Last time I went (two years ago) it was at the Google campus in Silicon Valley. That was cool, but it was great to get the opportunity to visit a new place. Munich is a beautiful city and has a lot to offer.
I had to be in Amsterdam the week before anyway, so I did not feel too bad about my carbon impact from flying to Europe. I did take the train from Amsterdam to Munich to save some carbon and more importantly: blog about my pretentiousness later. Along the same vein I got to the airport just now by Green Uber:
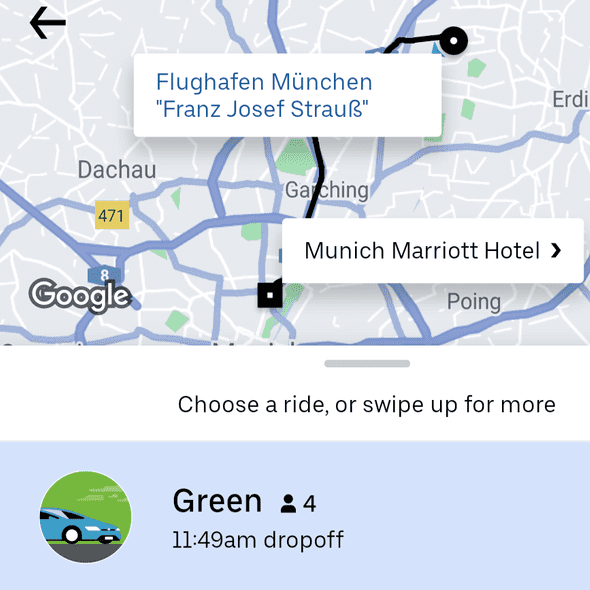
Haven’t seen this back home in New York yet. The price was actually slightly cheaper than Uber X. It was also my first time sitting in a Tesla Model 3. Not the best user experience trying to open the car door lol; I wasn’t able to get in or out of the car without the driver explaining to me how to operate the door. Munich had a bunch of other things I hadn’t seen in New York: electric scooters and bikes you can dump anywhere. Things do look slightly messy with those scooters and bikes scattered all over the city but it was pretty convenient. I didn’t end up using the subway at all, because the weather was perfect and it was so easy to rent scooters and bikes. In general I find the experience of navigating a new city so much easier compared to a decade ago, since you’re using all the apps and interfaces you’re familiar with from back home. That being said I did end up doing some stupid tourist things using the same apps. I took an electric scooter from Uber at night and Google Maps pointed me to go through this unlit park. Halfway through the park the scooter died on me. It was only then I realized that parking the scooter there would get me a 25 euro fine. I ended up trying to push the scooter out of the park before giving up and taking a cab back to the hotel.
Lightning Talks ⚡️
The next morning, decently well rested, Angelica and me presented at the Lightning Talks session. Organizations that signed up can tell a story about their students in under three minutes, see our two slides here:
The lightning talks are a great way to get a quick overview of all the different projects. My favorites were from Vicky Vergara from the Open Source Geospatial Foundation:
Watch the video @OSGeo
— vicky vergara (@VickyVvergara) September 6, 2019
@gsoc
2019 student Hang
working on pgRoutinghttps://t.co/llNxKPufXQ
and the Public Lab one presented by Jeffrey Warren:
I LOVE this slide from @jywarren - growing your community is what -gets- your work done, not something you do -instead- of work. @PublicLab encourages recent new contributors to prepare github issues for newbies - great onboarding path! #gsoc pic.twitter.com/08peKo7zEl
— yo yehudi🏳️🌈🇪🇺 (@yoyehudi) October 19, 2019
Unconference Sessions 🗣
Managing a Welcoming Open Source Organization
Since I enjoyed Jeffrey’s lightning talk so much, I decided to go to his session:
Amazing effort by @PublicLab to make #opensource contributing more inclusive: https://t.co/Ddz9Pu41lb. Great session by @jywarren at #GSOC. Feeling inspired!
— 🔧 Ino de Bruijn 🧬 (@inodb) October 20, 2019
I learned about the importance of language when writing contributing docs e.g.
"Please give back" vs "you have the ability to help others"
The former feels like a request whereas the latter emphasizes your unique abilities. For an example of that see code.publiclab.org:

Visual aids are very important as well for welcoming new contributors to your project. The GitHub checks give big red errors when contributors submit their pull request which can be discouraging:
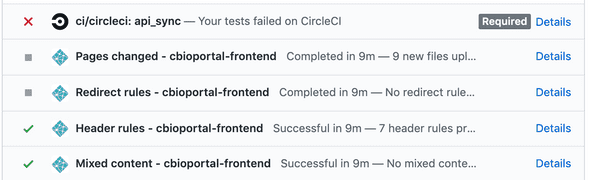
Showing this as a progress bar could be one way to make this less intimidating.
It was great to see in the session afterwards about Bots for maintainers and contributor onboarding organized by Oleg Nenashev that another major project like Jenkins made great efforts to incorporate similar ideas in their organization. Kai Blin remarked that people find it sometimes less intimidating to see a bot message something than him messaging the exact same thing himself. As an example he mentioned linting, e.g. a message like “improve the code styling here and here” was usually appreciated more when it came from a bot. Jeffrey added to this that in his organization they try to let encouraging words come from a human, since that is experienced as more welcoming. This leads to some interesting questions for an organization regarding what type of work should be done by bots versus humans. On the one hand it might be nice to automate parts of Pull Request reviews by bots but on the other hand the same task could be good for a relatively new contributor to the project so they feel good about contributing and for a new contributor to feel appreciated by a human being. I imagine metrics around contributor acquisition and what they end up contributing might help answer some of these questions.
Bioinformatics Related Sessions
There were a ton of other bioinformatics, life science and research/academics related organizations. Angelica and me organized a session at the start of the conference that was simply a round of introductions, which was helpful in getting acquainted with these types of orgs at GSoC. Angelica made great notes for our session: link. I ended up sitting down with Egon after. He organized a session around wikidata and I was curious about using their API for pathway data shown at wikipathways.org to feed the querying of cancer genomic data by pathway in cBioPortal:
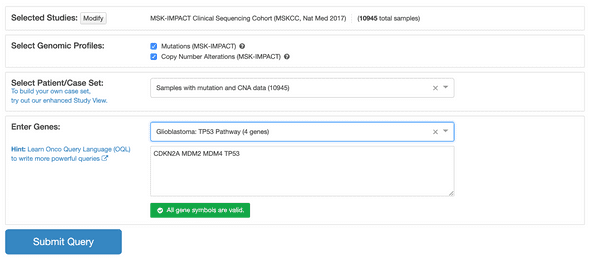
We actually had a GSoC project around integrating PathwayMapper into cBioPortal (more info here). Pathway mapper provides a great interface for people to curate their own pathways and subsequently query them in cBioPortal. Extending that with pathway data from wikidata seems like a great project. The neat thing is that wikidata connects all kinds of different entities with each other, allowing for instance to connect pathways to publications. That might provide for another interesting project to e.g. point users querying particular pathways on cBioPortal to relevant literature. I am on the wikipathways slack now so the first contact has been made 🙂
Collaborations with other organizations is something that came up during the session on universities as mentoring orgs. In our experience GSoC provides an excellent vehicle for collaborations between institutions. The pathway mapper integration project was a collaboration between our group and Bilkent University. The GDC import project was a first time collaboration between our group and the GDC team. This is a great way to involve more people in the open source process. I believe the academic world can learn a lot from the open source world. Open source development for instance often starts out in the open. There is a movement in the academic world to be more open, but it is usually only at time of publication. Somebody mentioned during the session that open source should probably be part of the curriculum. I did my undergrad in computer science and can’t remember any course that talked about open source and the process around contributing code. It might be different today but if not that should really change. Software carpentry is a great resource for educating scientists on this. For next year’s GSoC I’d like to try and involve more scientists. Another thing that Egon mentioned is that PhD students often list travel grants on their CV so having submitted a successful GSoC organization application is definitely something one can include there as well. Other ways to get academic credit for GSoC would be to publish papers on the work. We have successfully published on e.g. the CPTAC integration in cBioPortal and the G2S webservice. Another approach for open source tools is to list all contributors on the paper. Frequent publications of a tool can help give contributors academic credit.
These were just a few personal highlights of the unconference sessions. There is a full list of sessions and notes contributed by participants here:
https://docs.google.com/spreadsheets/d/18DwHPmqhh2rbxbbPA9OYitmlwsh3nI3azrngRuKfMGk/
TODO 🚧
I made a TODO list of all the things I want to follow up on after having been to the summit:
- Make an awesome contributing page like public lab: https://code.publiclab.org. Try to follow in their footsteps in how to make an inclusive community. Try using e.g. welcomebot. In addition to that it would be cool to adapt this a bit for our org. Like make a tutorial/demo on cBioPortal for people unfamiliar with cancer genomics and cancer. Our contributors are often a) people familiar with cancer genomics and little to no software engineering experience or b) people with software engineering experience but no cancer genomics knowledge. Some contributors might not be familiar yet with either. Figuring out some onboarding procedure for each of those would be great.
- Look into other open source programs such as Outreachy, specifically for underrepresented communities and Google Code In for high school students.
- Use wikidata to pull pathway data and incorporate in cBioPortal. If complicated this could be a good GSoC project for next year.
- Try to include more scientists/academics in GSoC. Try to convince them how it could be beneficial academically. Comentoring projects seems like a good approach. Need to start thinking about projects early.
- Some bots to try: welcomebot, releasedrafter, the CircleCI artifact bot to post links to our screenshot comparison page in the pull request. Currently it’s hard to find these for newcomers.
Goodbye and thank you! 👋
I furthermore had a bunch of other really great interactions with folks at GSoC. To name one: JJ Gao, our awesome team lead at cBioPortal receives a lot of notifications on GitHub but they don’t always end up in his inbox:
Just met https://t.co/bCRzWQBdKR at #GSoC - he gets all GitHub notifications intended for you @gaojj 😂😂😂 pic.twitter.com/UQaZfriZNo
— 🔧 Ino de Bruijn 🧬 (@inodb) October 19, 2019
A final thank you to Google, the organizers, the participants, the students and everyone that contributed to GSoC! Hope we will be able to participate again next year.
